Juniper Publishers-Open Access Journal of Case Studies
Whole Exome Sequencing Identifies Two Mutations in an Indian Patient with KBG Syndrome
Authored by Priyanka Vishwakarma
Abstract
KBG syndrome is a condition characterised by macrodontia, neurological disturbance, short stature, a distinct cranio-facial appearance, and also the skeletal anomalies. The authors describe what appears to be the first case of KBG syndrome with mutation in two different gene reported from the Indian subcontinent. Meticulous evaluation of the patients helps to classify such cases which may otherwise remain undiagnosed. Further research is reasonable to regulate the classic and the variant presentations of this condition, with the follow-up of the patients that provides the valuable data insights into its natural history and long-term prognosis. As per database these variants have been not reported till date, we found two variants in an Indian patient.
Keywords: KBG syndrome; Macrodontia; Short stature; Mental retardation
Introduction
KBG syndrome is a rare, genetic disorder that affects numerous body systems. There are around 200 patients reported till date with KBG syndrome and it has been described in the databanks or the research articles ever since year 1975 [1-16]. Till date the overall prevalence of the KBG syndrome has not been reported. The KBG syndrome was named after the initials of the last names of three original families reported by Herrmann et al. [17]. in 1975. Common clinical signs and symptoms in individuals with this disorder includes the unusual facial features, Short stature, skeletal abnormalities, and also intellectual disability, autistic features, sleep disturbances, feeding difficulties, hearing problems, speech delay, and learning difficulties. central upper incisors, cardiac defects, palate abnormalities, Short stature, skeletal abnormalities, and also intellectual disability, autistic features [12,18,19]. It is an autosomal dominant disorder including four principal appearances such as typical facial dysmorphism, macrodontia of the central maxillary incisors teethes, skeletal (mainly costovertebral) anomalies and the developmental delay [12]. However, this combination of features is observed in many genetic conditions. In addition, since many of the presenting features of KBG syndrome are mild and usually not associated with severe medical complications, it is likely to be undiagnosed. Hence, a complete understanding of the clinical features will be helpful in the diagnosis and the management of this syndromic condition. KBG syndrome may be suspected after a thorough clinical assessment, a detailed patient and family history, and the identification of characteristic physical findings. The diagnosis can also be made by gene panel analysis or next generation sequencing techniques, where multiple genetic causes of intellectual disability are investigated at the same time. Most Importantly it is caused by the pathogenic variants of ANKRD11 gene or by the chromosomal
Treatment of this condition is concentrating on the basis of particular specific clinical feature that are apparent in each affected cases [19]. In treatment standards requires the harmonized efforts of a team of specialists. On the basis if sign and symptoms the healthcare specialists may need to analytically and systematically plan an affected child’s treatment.
In such a condition the genetic counselling is acclaimed for the affected cases and their family members. The psychosocial support of the complete family may be beneficial as well [20].
The Orthopedic surgery is particularly very helpful to correct hip and spine or the skeletal abnormalities of the affected cases. Other devices such as hearing aids, speech therapy, and comprehensive dental care may also be advantageous. There are many cases where the children with KBG syndrome have been cured with growth hormone therapies. Initial results have been promising in helping children who are experiencing delays or diminished growth. More research is necessary to determine the long-term safety and effectiveness of growth hormone therapy in children with KBG syndrome [21,22].
Patients and Methods
In our Genetic Clinic, we took the blood from the patient and performed whole Exome Sequencing.
a) Patient 1: A 13-year-old boy, had presented to clinician with the slurred speech, short stature, microcephaly, dimorphism, jargon speech, mental retardation, intellectual disability-severe, continuous grimacing and abnormal EEG. With no family history (Table 1).
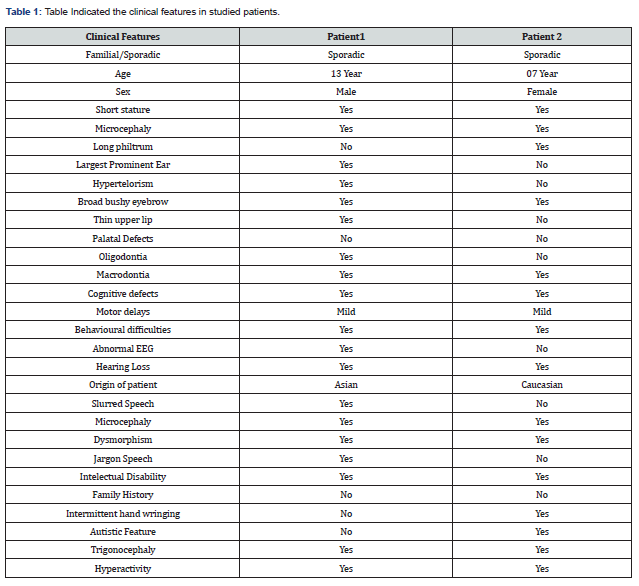
b) Patient 2: This patient is a 7-year-old girl, born to non-consanguineous parents, had presented to clinician with microcephaly, autistic like features & intermittent hand wringing with no family history (Table 1).
DNA extraction and quality and quantity measurements
Genomic DNA was extracted from 2mL peripheral venous blood samples of patient, with DNA extracted by using standard phenol–chloroform method [23]. The quality of DNA was assessed on 2% agarose gel electrophoresis, and quantity of DNA was measured by Nano Drop TM.
Whole exome sequencing
The DNA extracted from blood was used to perform the targeted gene capture using an Agilent sure select exome V6 kit. The libraries were sequenced to mean >100X coverage on Illumina sequencing platform and 6 to 8GB data will be generated. The sequences obtained are aligned to human reference genome (GRCh37/hg19) and variant analysis was performed using set of Bioinformatics Pipeline. Clinically relevant mutations were annotated using published variants in literature and a set of diseases databases – ClinVar, OMIM, GWAS, HGMD and SwissVar. Common variants are filtered based on allele frequency in 1000 Genome Phase 3, ExAC, EVS, dbSNP147, 1000 Japanese Genome etc. Non-synonymous variants effect is calculated using multiple algorithms such as PolyPhen-2, SIFT, MutationTaster2, Mutation Assessor and LRT. Only non-synonymous and splice site variants found in the clinical exome panel consisting of specific set of genes were used for clinical interpretation. Silent variations that do not result in any change in amino acid in the coding region are not reported.
Result
In patient 1 after performing whole exome sequencing a pathogenic variant (chr16:89337301 NM_001256183.1, c.7730C>G p. Ser2577Ter) has been identified in exon 12 of ANKRD11 gene with the heterozygous condition (Table 2) and also another variant (chr14:102478243 NM_001376.4, c.6650A>G p. Asn2217Ser) in the 33 exon of DYNC1H1 gene in heterozygous condition (Table 2). As per our and on the basis of databases these are not previously reported we are the who reporting this mutation from the Indian subcontinent. The variants claimed the clinical features also been correlated in this patient. Sanger validation has been done for this case to confirm the variant identified by Whole Exome Sequencing. Both parents were also tested and none of them was a carrier.
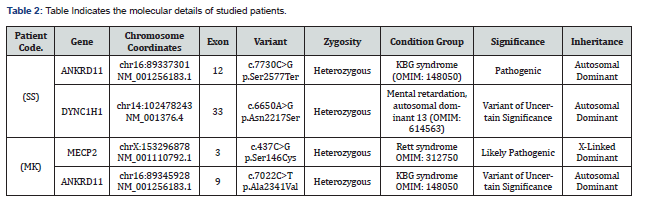
In patient 2 after performing whole exome sequencing two another variant we have identified a missense variant (c.7022C>T, p. Ala2341Val) (Table 2) in exon 9 of ANKRD11 gene and another missense variant (c.437C>G, p. Ser146Cys) (Table 2) in exon 3 of MECP2 gene. These both the variant has been reported already in databases. Both the variants are disease causing and has been correlated with patient’s clinical features. Patient showed the positive correlation with the clinical features. The sanger validation has been done in this patient to confirm the variant identified by Whole Exome Sequencing. Both parents were also tested and none of them was a carrier.
Discussion
In this study, we have identified a mutation of KBG syndrome – ANKRD11 (c.7730C>G, p. Ser2577Ter) Table 2. The common clinical features in the individuals with this condition includes infrequent facial features, skeletal deformities, and the intellectual debilities. A characteristic feature of the KBG syndrome is unusually large upper front teeth in other words the phenomena called macrodontia that has been observed in our both the patients. Other distinctive facial features include a wide, short skull (brachycephaly), a triangular face shape, widely spaced eyes (hypertelorism), wide eyebrows that may grow together in the middle (synophrys), a prominent nasal bridge, a long space between the nose and upper lip (long philtrum), and a thin upper lip. A common skeletal abnormality in people with KBG syndrome is slowed mineralization of the bones (delayed bone age) such as an affected 3-year-old child may have bones more typical of a child of 2. Including this the affected individuals can present the abnormalities of the bones of spine (vertebrae) and ribs. They can also have abnormalities of the bones of hands or feet, including unusually short or curved fifth (pinky or small) finger (brachydactyly or clinodactyly, respectively) and flat feet. Most of the affected individuals are shorter than the average height from birth. Growth of the mental health and the movement abilities is also delayed in this syndrome. Most affected individuals learn to speak and walk later than normal and have mild to moderate intellectual disability. Most people with this condition have behavioural or emotional problems, such as hyperactivity; anxiety; or autism spectrum disorder, which is characterized by impaired communication and social interactions.
Less common features of KBG syndrome comprise hearing loss, seizures, and the heart defects. Several ANKRD11 gene mutations have been establish to cause the KBG syndrome, a condition characterized by the large upper front teeth and other unusual facial features, skeletal abnormalities and intellectual disability. Most of these mutations lead to an abnormally short ANKRD11 protein, which likely has little or no function. Reduction of this protein’s function is thought to underlie the signs and symptoms of the condition. Because ANKRD11 is thought to play an important role in neurons and brain development, researchers speculate that a partial loss of its function may lead to developmental delay and intellectual disability in KBG syndrome. However, the mechanism is not fully identified. It is also unclear how loss of the ANKRD11 function leads to the skeletal features of the condition. Another type of mutation that affects the ANKRD11 gene, called 16q24.3 microdeletions, deletes genetic material from chromosome 16 in a region designated q24.3. The deleted region typically removes the ANKRD11 and ZNF778 genes, although other nearby genes may also be affected. People with this type of mutation have the similar clinical features to those affected with KBG syndrome, including unusual facial features and intellectual disability. Many also have brain abnormalities and features of autism spectrum disorders, which are characterized by impaired communication and socialization skills. Some researchers think that these microdeletions are different enough from KBG syndrome to be considered a separate disorder, called 16q24.3 microdeletion syndrome (Genetics Home Reference) this has been The dental dysmorphisms reported in patients with 16q24.3 microdeletions by Novara et al. [24].
Initially the molecular diagnosis of the KBG syndrome was not possible but since 2011 it has been possible, when the scientist Sirmaci and their colleagues [25]. acknowledged the deleterious intragenic heterozygous mutations in the gene responsible for KBG syndrome ANKRD11 in a group of patients with KBG syndrome concluded the technology whole genome sequencing. This gene makes the ankyrin repeat domain-containing protein 11, this protein is a member of the family of ankyrin repeat domain containing the cofactors that are the strongest inhibitors of ligand-dependent transcriptional activation. The ANKRD11 protein is restricted in the neurons and glial cells and it influences the expression of several genes linked to neural and bone growth. Mutations of this gene can arise surprisingly with absence of the family history or it can be inherited in an autosomal dominant manner. Microdeletion at 16q24.3 encompassing ANKRD11 or heterozygous mutation of ANKRD11 represents the common phenotype of KBG syndrome [26-30].
Ankyrin protein as its name suggests, this contains multiple regions called ankyrin domains; proteins with these domains help other proteins interact with each other [31]. This protein interacts with certain proteins called the histone deacetylases, which are important for controlling gene activity. Through these interactions, ANKRD11 affects when genes are turned on and off. For example, ANKRD11 brings together histone deacetylases and other proteins called p160 coactivators [32]. This association regulates the ability of p160 coactivators to turn on gene activity. ANKRD11 may also enhance the activity of a protein called p53, which controls the growth and division (proliferation) and the self-destruction (apoptosis) of cells. The ANKRD11 protein is found in nerve cells (neurons) in the brain [33]. During embryonic development, ANKRD11 helps regulate the proliferation of these cells and development of the brain. Researchers speculate that the protein may also be involved in the ability of neurons to change and adapt over time (plasticity), which is important for learning and memory [34]. ANKRD11 may function in other cells in the body and appears to be involved in normal bone development (Genetics Home Reference).
In patient 1, we found one copy (heterozygous) of a nonsense variant (c.7730C>G, p. Ser2577Ter) in exon 12 of ANKRD11 gene (Table 2). Identified variant predicted to produce the truncated protein. Based on the literatures and in silico prediction tools scores, particular variant is disease causing by Mutation Taster. As per our knowledge and on the basis of literature survey and databases the variant is pathogenic, it has been not reported previously. This variant is also relevant with the clinical phenotype of the individual, but has to carefully correlate with clinical symptoms or parental studies (inherited or de novo). With accordance to ACMG guidelines and literatures, this particular variant is classified as pathogenic.
Another variant in this patient 1 is, found in exon 33 of DYNC1H1 gene with variant (c.6650A>G, p. Asn2217Ser) (Table 2) associated with the Mental retardation, autosomal dominant 13. MRD13 is an autosomal dominant form of mental retardation associated with variable neuronal migration defects resulting in cortical malformations. More variable features include earlyonset seizures and mild dysmorphic features. Some patients may also show signs of peripheral neuropathy, such as abnormal gait, hyporeflexia, and foot deformities (OMIM: 614563) [35,36]. DYNC1H1 gene is responsible for instructions for making a protein that is part of a group (complex) of proteins called dynein. This complex is found in the fluid inside cells (cytoplasm). Dynein is turned on (activated) by attaching (binding) to another complex called dynactin. This dynein-dynactin complex binds to various materials within cells. Using energy provided by molecules called ATP, the dynein-dynactin complex moves material along a track-like system of small tubes called microtubules, similar to a conveyer belt. The dyneindynactin complex is necessary for protein transport, positioning of cell compartments, movement of structures within the cell, and many other cell processes. Dynein helps neighbouring nerve cells (neurons) communicate by transporting sac-like structures called synaptic vesicles that contain chemical messengers. When synaptic vesicles are passed from one neuron to another, the dynein-dynactin complex transports the vesicle from the edge of the cell to the nucleus, where the chemical message is received.
The parts (subunits) of a dynein complex are classified by weight as heavy, intermediate, light intermediate or light chains. Two heavy chain proteins bind together to form the core of the dynein complex. Combinations of intermediate, light intermediate and light chains make up the rest of the complex. The protein produced from the DYNC1H1 gene is a heavy chain. Other subunits are produced from different genes (Genetics Home Reference).
Based on the literatures and in silico prediction tools scores particular variant is disease causing by Mutation Taster. This variant may be relevant to clinical phenotype of the individual but the clinical significance is largely unknown. Thus, this particular variant is classified as variant of uncertain significance (VUS). Hence, this variant has to carefully correlate with clinical symptoms or parental studies (inherited or de novo).
Patient 2 carries one copy (heterozygous) of a missense variant(c.7022C>T, p. Ala2341Val) (Table 2) in exon 9 of ANKRD11 gene. The identified variant affects the protein functioning. On the basis of literatures and in silico prediction tools scores, particular variant is pathogenic or disease causing by Mutation taster & possibly damaging by PolyPhen. This variant has been reported in dbSNP database with identification number of rs4031012 and in gnomAD database with an allele frequency as 0.000009488. This variant is also relevant to the clinical phenotype of the patient but the clinical significance is of this variant is not much clinically relevant or largely unknown. Thus, this particular variant is classified under the category of variant of uncertain significance (VUS). Hence, this variant required to carefully correlate with clinical symptoms or parental studies (inherited or de novo).
In this patient 2, we have found another variant carries one copy (heterozygous) of a missense variant (c.437C>G, p. Ser146Cys) (Table 2) in exon 3 of MECP2 gene. This variant affects the functioning of the protein. Based on the literatures and in silico prediction tools scores, particular variant is pathogenic or disease causing by the Mutation taster and tolerated by SIFT. The variant has been well reported in dbSNP database with identification number of rs61748390 and in ClinVar database with ID number: VCV000143562.2 as Pathogenic/Likely pathogenic. Thus, this particular variant is classified as Likely Pathogenic or likely to disease causing.
MECP2 gene mutation that has been identified (c.437C>G, p. Ser146Cys) (Table 2) is responsible for the Rett syndrome. It’s a brain disorder that happens almost exclusively in girls. The most common form of the condition is known as classic Rett syndrome. After birth, girls with classic Rett syndrome have 6 to 18 months of apparently normal development before developing severe problems with language and communication, learning, coordination, and other brain functions. Early in childhood, affected girls lose purposeful use of their hands and begin making repeated hand wringing, washing, or clapping motions. They tend to grow more slowly than other children and about threequarters have a small head size (microcephaly). Other signs and symptoms that can develop include breathing abnormalities, spitting or drooling, unusual eye movements such as intense staring or excessive blinking, cold hands and feet, irritability, sleep disturbances, seizures, and an abnormal side-to-side curvature of the spine (scoliosis) (Genetic Home Reference).
Mutation in MECP2 gene that provides instructions for making a protein called MeCP2. This protein helps regulate gene expression activity by modifying the chromatin, the complex of DNA and protein that packages DNA into the chromosomes. The MeCP2 protein is present in throughout the cells of the body, though it is predominantly abundant in brain cells. In the brain, this protein plays an important role for the utility of several types of cells, such as nerve cells (neurons). The protein likely plays a role in maintaining connections (synapses) between the neurons, where cell to-cell communication occurs. Many of the genes that are known to be regulated by the MeCP2 protein also plays a very important role in normal brain function, particularly for the maintenance of synapsis processes.
Scientists have the confidence that the MeCP2 protein also involved in processing molecules called messenger RNA (mRNA), which serve as genetic blueprints for making proteins. By cutting and rearranging mRNA molecules in different ways, the MeCP2 protein controls the production of different versions of certain proteins. This process is known as alternative splicing. In the brain, the alternative splicing of proteins is critical for normal communication between neurons and may also be necessary for the function of other types of brain cells (Genetic Home Reference).
Conclusion
In conclusion this study showed the utility of Whole exome sequencing in misdiagnosed pathogenic cases. By this study we reported two mutations in the Indian subcontinent. Whole exome approach provides a major advance in likely diagnostic yield in suspected inherited mutation containing cases, this methodology may uncover unexpected findings such as mutations in genes known to cause familial.
To know more about Juniper Publishers please click on: https://juniperpublishers.com/manuscript-guidelines.php
For more articles in Open Access Journal of Case Studies please click on: https://juniperpublishers.com/jojcs/index.php


